Key-Words- V. cholerae O1, El Tor, MAMA PCR, ctxB
INTRODUCTION- The acute diarrheal disease cholera is caused by Vibrio cholerae, a major public health problem in Asia, Africa and Latin America1. According to the World Health Organisation (WHO) estimates there were 3-5 million cholera cases and 1,00,000- 1,20,000 deaths each year throughout the world2. V. cholerae has more than 200 known serogroups but not all the strains are pathogenic. Among them, O1 and O139 antigens are highly patho-genic and acknowledged to cause epidemic and pandemic disease3. The symptom of the acute cholera is rice water stools.Clinical manifestations of the disease range from mild symptoms such as abdominal cramps, nausea and vomiting, to more severe symptoms such as dehydration, shock and death4. A devastating cholera outbreak, that had occurred in Haiti since October 2010, involved 6, 98, 893 cases and 8,540 deaths5. V. cholerae serogroup O1 is classified into two biotypes, which are termed ‘classical’ and ‘El Tor’6. The current seventh pandemic of cholera is caused by the El Tor biotype was originated in 1961 from Celebes Islands in Indonesia7. Spread of this biotype was recorded for the first time during 1964 in India and it made its first appearance in Delhi during June, 19658. In recent years, it has been seen that the emergence of new variants of V. cholerae O1 have carried characteristics of both classical and El Tor biotypes and these variants of V. cholerae O1 are called as ‘atypical El Tor’. These strains might have evolved from El Tor variants that acquired certain characteristics from classical genome9. In this study, we focused on type of CT genotype existing among V. cholerae O1 isolated from the patients who were admitted in MVIDH, Delhi during 2012-2014.
MATERIALS AND METHODS-
Collection and processing of samples- 30 V. cholerae O1 isolates were revived from stock cultures were maintained at Laboratory Department in MVIDH, Delhi during 2012-2014. These samples were revived and transferred to alkaline peptone water (APW, pH-8.6) and incubated at 37°C for 4-6 hrs. After incubation, the enriched cultures were further inoculated to the thiosulphate-citrate-bile-salts-sucrose (TCBS) agar and bile salt agar (BSA, pH-8.6) (Hi-Media, Mumbai)10.
Confirmation of V. cholerae- Typical colonies appearing on TCBS/BSA were con-firmed by standard biochemical tests10. These isolates were further tested serologically11 with commercially available V. cholerae O1 polyvalent and monovalent and O139 antiserum (BD, USA and Denka Seiken, Ltd. Japan).
Genomic DNA preparation- Genomic DNA from each isolate was extracted using protocol described previously12.
PCR assays for detection of ctxB genotype- Mismatch amplification mutation assay was used for classifying the strains into prototype El Tor, hybrid, or El Tor variant biotype based on their ctxB gene13. In this PCR, the common forward primer for both classical and El Tor alleles was used. Two allele specific primers Re-Cla and Re-Elt were used respectively. The PCR conditions and cycles were described previously13. V. cholerae O1 strains 569B (classical) and N16961 (El Tor) were used as control strains. The primers used were synthesized by Invitrogen, India and dNTPs, Taq polymerase, 10X Taq buffer used in this study were obtained from Genei, Bangalore. The primer sequences were used in this PCR is given in Table 2.
RESULTS-
Characterisation of isolates- A total of 30 V. cholerae O1 isolates were revived from maintained isolates during 2012-2014 (Table 1). All isolates were biochemically confirmed as V. cholerae biotype El Tor and serologically confirmed with polyvalent O1 and then agglutination with monovalent Ogawa.
Table-1. Showing types of ctxB in Delhi isolates during 2012-2014
| No. of isolates | Year | Serogroup/Serotype | ctxB |
|---|---|---|---|
| 10 | 2012 | O1/Ogawa | Classical |
| 10 | 2013 | O1/Ogawa | Classical |
| 10 | 2014 | O1/Ogawa | Classical |
Table -2. Primer sequences were used in MAMA PCR
| Gene (S) | Primer Sequence | Amplicon Size (bp) | Ref. |
|---|---|---|---|
| ctxB FW | ACTATCTTCAGCATATGCACATGG | 13 | |
| Re-El | CTGGTACTTCTACTTGAAACA | 186bp | 13 |
| Re-Cla | CTGGTACTTCTACTTGAAACG | 186bp | 13 |
Confirmation of ctxB gene by MAMA PCR- All 30 isolates were screened for ctxB gene by MAMA PCR with two allele specific primers, one for Classical and other for El Tor. All isolates were amplified with classical specific primers (Fig. 1). Only control strain N16961 were amplified with El Tor specific primers (Fig. 2).
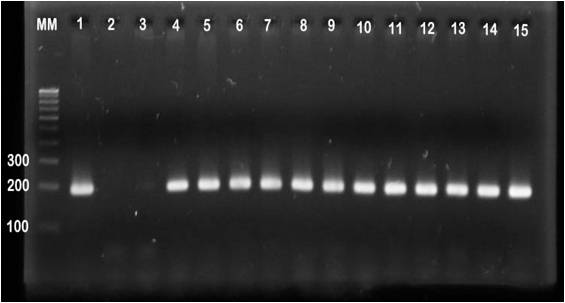
Figure 1: MAMA PCR assay of agarose gel electrophoresis of ctxB yielded 186bp amplicon size fragment when using primer pair FW-Com with Re-Cla. Lane M, 100bp molecular weight marker, lane1-569B Positive Control, lane 2- N16961 Control strain for El Tor, lane 3- Negative Control and lane 4-15 test strains shown positive band
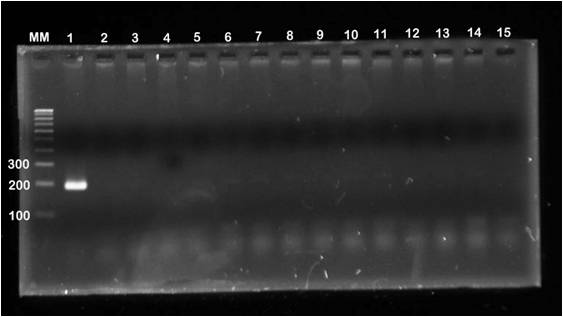
Figure 2: MAMA PCR assay of agarose gel electrophoresis of ctxB yielded 186bp amplicon size fragment when using primer pair FW-Com with Re-Elt. Lane M, 100bp molecular weight marker, lane1-N16961 Positive Control, lane 2- 569B Control strain for Classical, lane 3- Negative Control and 4-15 test strains shown negative band
CONCLUSION- In the present study, all isolates are phenotypically El Tor but carried ctxB gene of classical biotype and known as altered El Tor. Altered El Tor is more patho-genic than El Tor because it harboured ctxB gene of classical biotype and better survival in the environment. More molecular study is needed to understand the changing epidemiology of V. cholerae O1 infections and better planning to control cholera disease in this part of the country.
REFERENCES
- Farque SM and Nair GB (2002). Molecular Ecology of Toxigenic Vibrio Cholerae. Microbiol Immunol 46: 59-66.
- Taylor DL, Kahawita TM, Cairncross S, Ensink JHJ (2015) The Impact of Water, Sanitation and Hygiene Interventions to Control Cholera: A system Review. PLoS ONE 10: 1-19, DOI:10.1371/journal.pone.0135676.
- Reidl J and Klose KE (2002) Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26: 125-139.
- Weir E and Haider S (2004) Cholera outbreaks continue. CMAJ 170: 1092-1093.
- UN FACT SHEET (2014) Combatting Cholera in Haiti. www.un.org/news/dh/infocus/haiti/Cholera_UN_Factsheet_24%20Feb_2014. Accessed on 24.6.2015.
- Samadi AR, Huq MI, Shahid N, Khan MU, Eusof A, Rahman AS et al., (1983) Classical Vibrio cholerae biotype displaces EL tor in Bangladesh. Lancet 1: 805-807.
- Faruque SM, Albert MJ, and Mekalanos JJ (1998) Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62: 1301-1314.
- Singh J, Bora D, Sharma RS, Khanna KK, Verghese T (1995) Epidemiology of cholera in Delhi-1992. J Trop Pediatr 4l: 139-42.
- Safa A, Nair GB and Kong RYC (2009) Evolution of new variants of Vibrio cholerae O1. Trends in Microbiol 18: 46-53.
- World Health Organization (2003) Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. Centre for Disease Control and Prevention, Atlanta Georgia USA. USAID/WHO/CDC 141-159.
- Kay BA, Bopp CA, Wells JG (1994).Isolation and identification of Vibrio Cholerae O1 from fecal specimens. In: Wachsmuth Ik, Blake PA, Olsvik O, editors. Vibrio cholerae and cholera: Molecular to global perspectives. Wahington DC, USA: ASM Press; p.3-25.
- Singh P, Kumar D, Prasad Y, Ramamurthy T, Sarkar BL, Sharma NC (2015). Prevalence of multidrug resistant altered Vibrio cholerae O1 isolates among Diarrhoeal patients in Delhi during 2008-2012. Indian J Appl Res 5: 624-628.
- Morita M, Ohnishi M, Arakawa E, Bhuiyan NA, Nusrin S, Alam M et al., (2008) Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol 52:314-317.
- Ramamurthy T and Sharma NC (2014) Cholera Outbreaks in India. Current Topics in Microbiology and Immunology 379: 49-85.
- Kanungo S, Sah BK, Lopez AL, Sung JS, Paisly AM, et al. (2010) Cholera in India: an analysis of reports, 1997-2006. Bull World Health Organ 88: 185-191.
- Kaper JB, Morris JG Jr., and Levine MM (1995) Cholera. Clin Microbiol Rev 8: 48-86.
- Sack DA, Sack RB, Nair GB, Siddique AK (2004) Cholera. Lancet 363: 223-233.
- Nair GB, Qadir F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA et al., (2006) Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44: 4211-3.
| Source of Financial Support: Nil Conflict of interest: Nil |
| International Journal of Life-Sciences Scientific Research (IJLSSR) Open Access Policy Authors/Contributors are responsible for originality, contents, correct references, and ethical issues. IJLSSR publishes all articles under Creative Commons Attribution- Non-Commercial 4.0 International License (CC BY-NC). https://creativecommons.org/licenses/by-nc/4.0/ |